Linear Regression on RDKit fingerprints¶
Here is a simple prediction of water solubility using the FreeSolv database. The solubility is defined as a log solubility in moles per litre.
I apply linear regression to Morgan fingerprints. By default, splitting uses 75% of the dataset for training, with the remainder reserved for testing.
import pandas as pd
from rdkit import Chem
from rdkit.Chem import AllChem
from sklearn.linear_model import LinearRegression
from sklearn.model_selection import train_test_split
df = pd.read_csv("https://deepchemdata.s3-us-west-1.amazonaws.com/datasets/SAMPL.csv")
fpgen = AllChem.GetMorganGenerator(radius=3, fpSize=2048)
# add a column with fingerprints
df["fps"] = df["smiles"].apply(
lambda smiles: fpgen.GetFingerprintAsNumPy(Chem.MolFromSmiles(smiles))
)
train, test = train_test_split(df)
reg = LinearRegression().fit(train.fps.to_list(), train.expt)
score = reg.score(test.fps.to_list(), test.expt)
Score¶
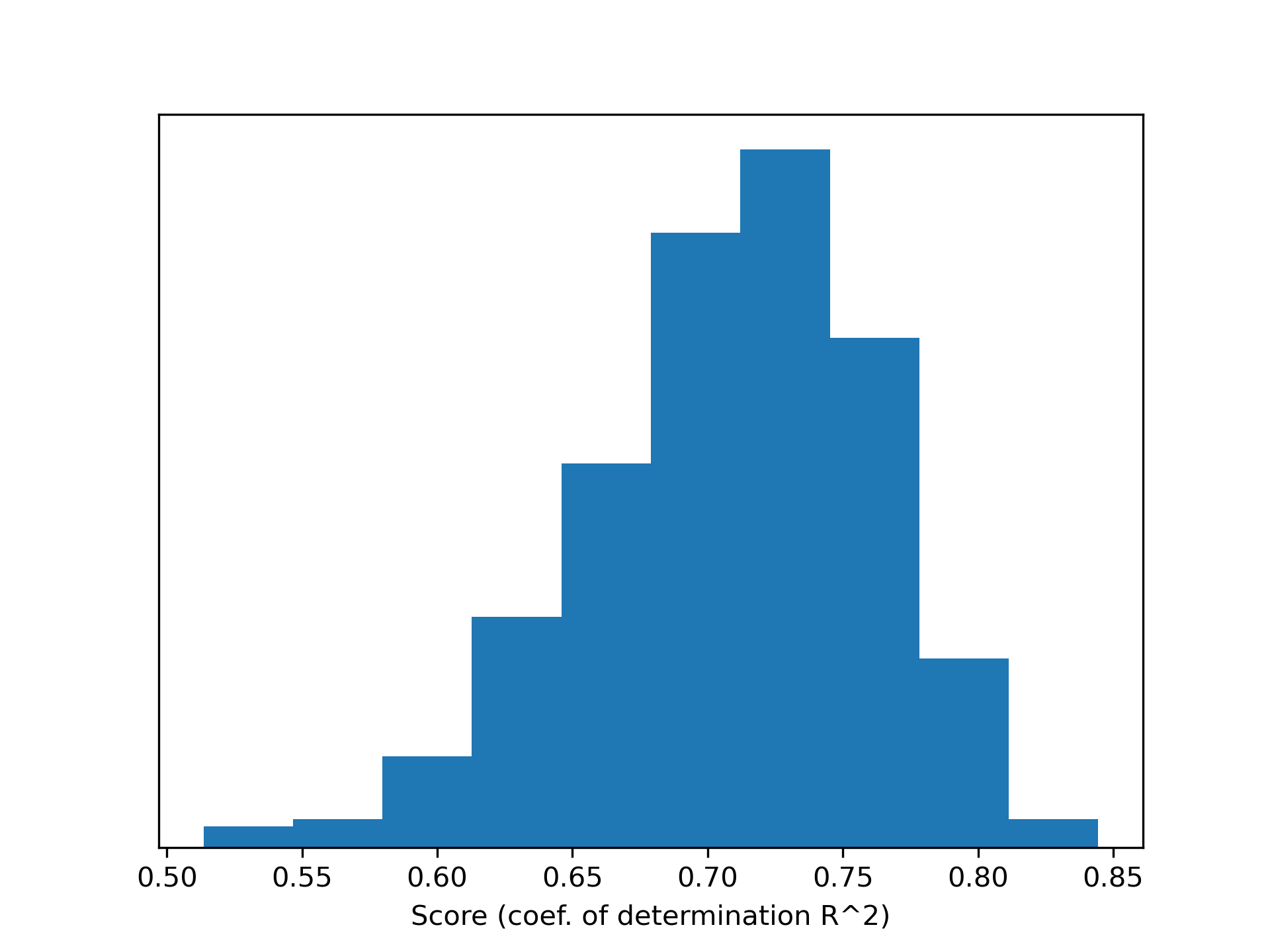
The average score is typically between 0.70 and 0.75, while the best possible value is 1. Specifically, the score is defined as the coefficient of determination:
When every prediction is perfect, the numberator becomes 0, so \(R^2\) reaches 1.
It is interesting to investigate the impact
of different splits on performance.
Depending on which part of the dataset
is used for training and testing, results can vary significantly:
Pandas, NumPy and scikit-learn intersection¶
Although this code is straightforward, there's an interesting trap: I used Pandas DataFrame and stored the fingerprints as "cells", ideally letting us do:
However, this fails because the fingerprints add another dimension. The.to_numpy() call returns a one-dimensional array
of objects (shape = (rows_n, )) instead of the expected
two-dimensional shape ((rows_n, 2048)). That leads to a conversion error.
One workaround is to use .to_list(), which preserves each
row’s fingerprint as a separate array that scikit-learn
correctly interprets as 2048-dimensional.
So watch out for dimensional mismatches when
using fingerprints in scikit-learn.